Abstract
Differential expression profiling has identified specific genes and pathways that are variably expressed in different subsets of acute myeloid leukemia. However, comprehensive evaluation of genes and pathways whose expressions are uniquely altered in AML compared to that in normal hematopoiesis is lacking. Previously we used whole transcriptome sequencing (RNA Seq) to investigate the expression profiles of t(8;21) and Inv(16) translocations, defining that down-regulation of homeobox (HOX) gene family plays an important role in CBF (core binding factors)-AML hematopoiesis compared to other AML variants. In this study, we used RNA seq data from diagnostic specimens obtained from 473 children and young adults with AML to compare their transcriptome to that in normal bone marrow (NBM) from healthy individuals (N=20).
Initial comparison of AML vs. NBM transcriptome identified 2,151 genes to be differentially expressed with 2-fold change and FDR <0.01 using GLM (Generalized Linear Models) analysis; the most up/down-regulated genes are DUSP10 (1.06E-42), FCRL6 (5.11E-146) PLIN3 (2.19E-28) and TTC16 (22.46E-124); GSEA (Gene Set Enrichment Analysis) showed that these 2,151 genes were mostly involved in "p53 signaling pathway" (hsa04115; FDR=0.0013), "regulation of T cell mediated cytotoxicity" (GO:0001914; FDR=0) and "decreased circulating interleukin-17 level" (MP:0008615; FDR=0).
In an additional analysis, in order to more appropriately capture transcripts whose expression might be limited to a small subset of patients, we used a customized filtering approach (RPKM value >5 in at least 10% AML and <5 in at least 80% normal samples) to maximize our ability to identify genes that are uniquely expressed or silenced in AML vs. NBM. This analysis identified 1,173 genes to be most AML-specific with the customized cutoffs. This analysis identified expression of a number of Cancer Testis Antigen (CTA) family of genes including PRAME, and CCNA1 that were highly expressed in a significant subset of the patients. GSEA top terms for these AML-specific genes were "mRNA surveillance pathway" (hsa03015; FDR=0.0024), "autophagosome assembly" (GO:0000045; FDR=0.0027) and "increased circulating interleukin-13 level" (MP:0008608; FDR=0).
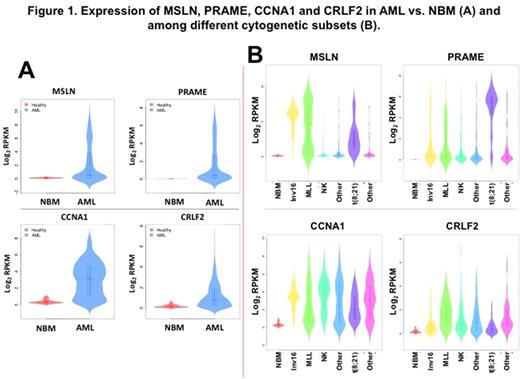
There were 261 genes identified by both methods, including MSLN (Mesothelin), PRAME (Melanoma antigen preferentially expressed in tumors) and CCNA1 (Cyclin-A1) (Figure 1A). GSEA of these 261 genes showed that the most significant GO pathway was "negative regulation of transcription from RNA polymerase II promoter" (GO:0000122; FDR=0.0025).
We also applied the same pipeline for the comparisons between NBM with each AML cytogenetic subgroup (Inv(16), t(8:21), MLL and normal karyotype(NK)), each comparison resulted in 2,000~3,000 DE (differentially expressed) genes from GLM, with t(8;21) vs. NBM the largest number (2,318 genes). There were also 200~400 overlapped genes identified by both GLM and cutoff approaches; For GSEA results, "MicroRNAs in cancer" (hsa05206; FDR=0.0017) and "NF-kappa B signaling pathway" (hsa04064; FDR=0.0018) were shown in Inv(16) vs. NBMs; "regulation of transcription involved in G1/S transition of mitotic cell cycle" (GO:0000083; FDR= 0.0015) was shown in t(8;21) vs. NBM; "platelet degranulation" (GO:0002576; FDR= 0.0021) was shown in MLL vs. NBM; "NOD-like receptor signaling pathway" (hsa04621; FDR=0.0023) was shown in NK vs. NBM.
MSLN and PRAME displayed distinct expression patterns among different cytogenetic groups: MSLN was highly expressed in most inv(16) samples; however, PRAME was highly expressed in the majority of t(8;21) samples. Moreover, we grouped all samples (N=493) based on the expression of MSLN (RPKM cutoff=5) or PRAME (RPKM cutoff=4); and performed the same analysis using GLM and GSEA (Figure 1B). 164 differentially expressed genes were identified, and the most significant GO pathway was "platelet degranulation" (GO:0002576; FDR=0.00057) in MSLN-based groups; 186 genes were identified, and "G-protein coupled receptor binding" (GO:0001664; FDR=0.0024) was the most significant one in PRAME-based groups.
This analysis provides a comprehensive gene expression profile comparison between AML and normal hematopoiesis, incorporating current knowledge of pathways/interactions from databases, especially for those of clinical interests/significance.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.